PICAchooser (the program)
Basically, PICAchooser loads the results of a MELODIC ICA run and lets you step through the components, look at them, and decide if you want to keep them or discard them. After you load the dataset, you get a window showing the active IC component (both the timecourse and the spatial map), the power spectrum of the active IC component, and the motion timecourses for comparison. By default, components are flagged to be kept (and the timecourses are in green*).
Usage
usage: PICAchooser runmode [options]
A program to review (and alter) melodic component selections.
- positional arguments:
- runmode Analysis mode. Valid choices are “melodic”,
“groupmelodic” “aroma”, and “fix”. In melodic mode, the default output file is named “badcomponents.txt” and will be written to MELODICDIR as comma separated integers. In groupmelodic mode, the default output file is named “goodcomponents.txt” (you are more interested in which components should be retained) and will be written to MELODICDIR as newline separated integers (starting from 0). In aroma mode, the file “classified_motion_ICs.txt” must exist in the parent of MELODICDIR; by default the output will be written to “classified_motion_ICs_revised.txt” in the same directory. In fix mode, the default output file is named “hand_labels_noise.txt” and will be written to MELODICDIR as comma separated integers with square brackets surrounding the line.
- optional arguments:
- -h, --help
show this help message and exit
Standard input file location specification. For certain runmodes, one of these will be sufficient to fully specify all file locations.:
- --featdir FEATDIR
The FEAT directory associated with this MELODIC run.
- --melodicdir MELODICDIR
The .ica directory for this MELODIC run.
Nonstandard input file location specification. Setting any of these overrides any location inferred from –melodicdir or –featdir.:
- --backgroundfile BGFILE
The anatomic file on which to display the ICs (usually found in FEATDIR/reg/example_func.nii.gz),
- --funcfile FUNCFILE
The functional file to be filtered (usually found in FEATDIR/filtered_func_data.nii.gz),
- --motionfile MOTIONFILE
The anatomic file on which to display the ICs (usually found in FEATDIR/mc/prefiltered_func_data_mcf.par). If the file has a .tsv extension, assume it is an fmriprep confounds file.
- --ICfile ICFILE
The independent component file produced by MELODIC (usually found in MELODICDIR/melodic_IC.nii.gz).
- --ICmask ICMASK
The independent component mask file produced by MELODIC (usually found in MELODICDIR/mask.nii.gz).
- --timecoursefile MIXFILE
The timecourses of the independant components (usually found in MELODICDIR/melodic_mix),
- --ICstatsfile STATSFILE
The melodic stats file (usually found in MELODICDIR/melodic_ICstats),
- Other arguments:
- --initfile INITFILE
The name of an initial bad component file (in aroma mode, this overrides the default input file for AROMA).
- --outputfile OUTPUTFILE
Where to write the bad component file (this overrides the default output file name).
- --filteredfile FILTEREDFILE
The name of the filtered NIFTI file. If this is set, then when the bad component file is written, the command to generate the filtered file will be printed to the terminal window.
- --displaythresh DISPLAYTHRESH
z threshold for the displayed ICA components. Default is 2.3.
- –spatialroi XMIN XMAX YMIN YMAX ZMIN ZMAX
Only read in image data within the specified ROI. Set MAX to -1 to go to the end of that dimension.
- Configuration arguments:
- --keepcolor KEEPCOLOR
Set the color of timecourses to be kept (default is “g”).
- --discardcolor DISCARDCOLOR
Set the color of timecourses to discard (default is “r”).
- –transmotlimits LOWERLIM UPPERLIM
Override the “normal” limits of translational motion from the values in the configuration file to LOWERLIM-UPPERLIM mm.
- –rotmotlimits LOWERLIM UPPERLIM
Override the “normal” limits of rotations motion from the values in the configuration file to LOWERLIM-UPPERLIM radians.
- --scalemotiontodata
Scale motion plots to the motion timecourse values rather than to the limit lines.
- --componentlinewidth LINEWIDTH
Override the component line width (in pixels) in the configuration file with LINEWIDTH.
- --motionlinewidth LINEWIDTH
Override the motion timecourse line widths (in pixels) in the configuration file with LINEWIDTH.
- --motionlimitlinewidth LINEWIDTH
Override the line widths of the motion limit lines (in pixels) in the configuration file with LINEWIDTH.
- Debugging arguments:
- --verbose
Output exhaustive amounts of information about the internal workings of PICAchooser. You almost certainly don’t want this.
Example
Run PICAchooser to look at a series of independent components and assign them a rating:
PICAchooser RUNMODE --featdir FEATDIRECTORY --melodicdir MELODICDIRECTORY
You’ll then get a window that looks like this:
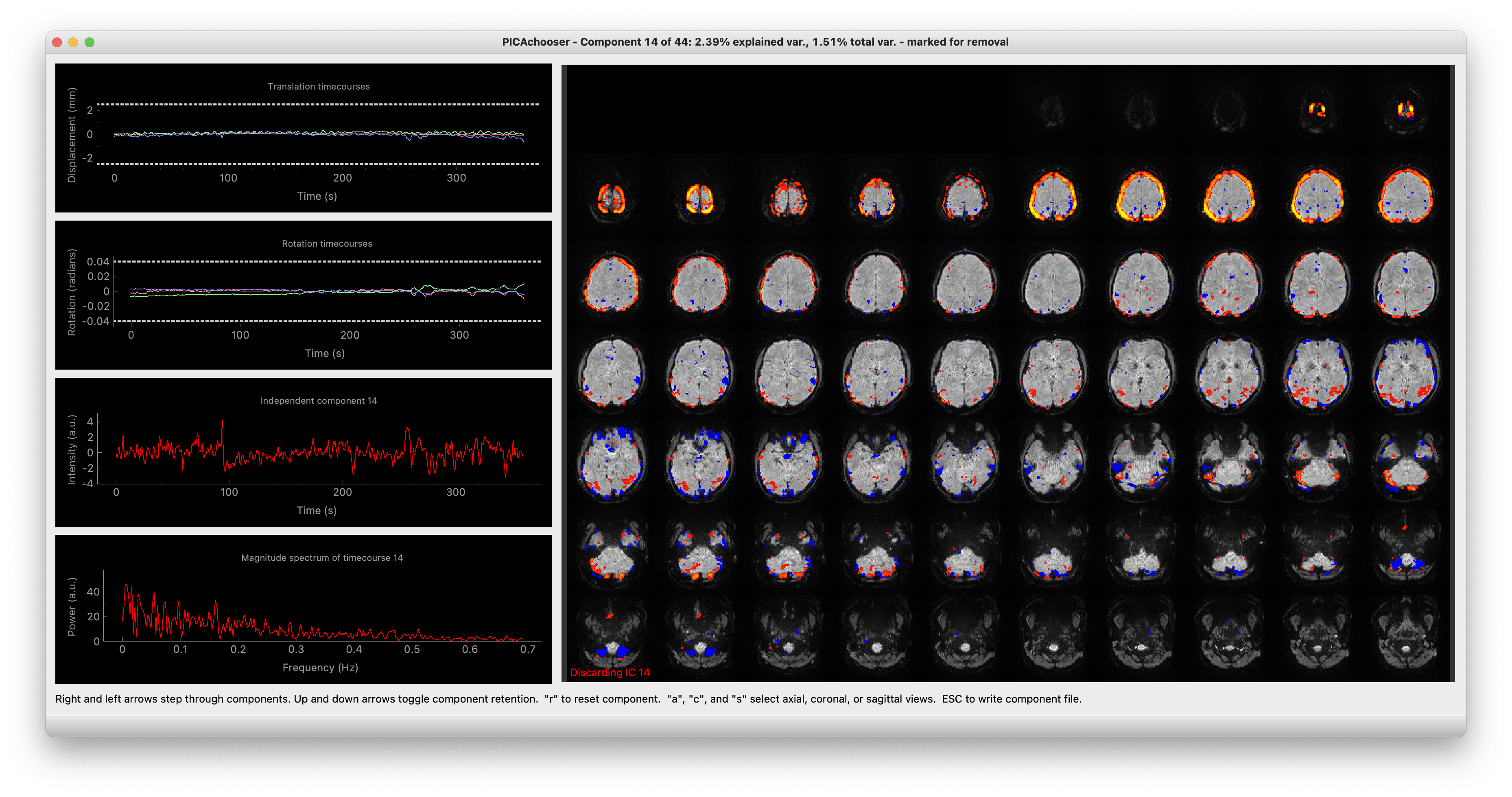
PICAchooser screenshot
Controls
To toggle whether the current component should be kept or discarded, press the up or down arrow key. You can change back and forth as much as you want. Components to be discarded are in red, ones to be kept are in green*.
To go to the next (or previous) component, press the right (or left) arrow. You’ll wrap around if you hit the end.
Press the escape key at any time to save the current version of the component list. The component list is saved automatically when you quit.
Input file specification
For most datasets, you only need to specify the FEAT directory where the preprocessing was done, and the MELODIC directory where the ICA analysis was performed, and PICAchooser can find all the files it needs to let you do component selection. In some cases, however (looking at you, fmriprep), the files you need to find can be scattered all over the place, with different names (and even different formats). In those cases, you can specify the name and location of every one of the files separately (anything you set with these options will override the default locations calculated from the FEATDIR and MELODICDIR).
Other command line options
--initfile lets you read in a bad component file from anywhere to
use as a starting point in your classification. It’s the normal behavior
in aroma mode (reading from MELODICDIR/../classified_motion_ICs.txt),
but you can do it in any mode with this flag, and it will override the
aroma classifications.
--outputfile lets you write the bad component file anywhere you
want, rather than just the default location.
--filteredfile specifies where the filtered file would go. If this
is set, PICAchooser prints the fsl_regfilt command to filter the data
using the currently tagged bad components whenever the file is saved
(when the escape key is pressed, or when you quit).
--displaythresh sets the z-threshold for the component maps.
--spatialroi XMIN XMAX YMIN YMAX ZMIN ZMAX lets you zoom in on a
cubic ROI within the NIFTI dataset. Useful if you did a constrained ICA
on a particular brain region. Set the MAX value to -1 to go to the
maximum value for a given dimension. These are voxel indices, with 0
being the first element of each dimension.
* Configuration changes
You can use --keepcolor, --discardcolor, --transmotlimits
and --rotmotlimits to alter display behavior for the current run
(useful if you’re using the docker container). To change things
semi-permanently, edit the file ${HOME}/.picachooser.json. This file is
created with default values if it is not present. You can use any valid
python color specification string for color values, e.g. “r”, “ff0000”,
or “FF0000” could all be used for red.
--componentlinewidth, --motionlinewidth, and
--motionlimitlinewidth can all be used to specify various linewidths
(in pixels) for the various plots. Useful if you want to make a
screenshot pretty for a figure.
--scalemotiontodata autoscales the motion plots to the motion
timecourse values rather than to fixed limits.
The motion plots have two dotted lines to indicate “normal” motion limits (by default +/-2.5 mm for translation and +/-0.04 radians for rotation). The locations of these lines are set by “transmotlimits” and “rotmotlimits” in the configuration file. Setting “motionplotstyle” to 0 will remove the lines, and fix the y range of the plots to the limit values. Set the limit line color using “motionlimitcolor”.
Outputs
In melodic mode, the default output file is named “badcomponents.txt”; components flagged for removal will be written to MELODICDIR as comma separated integers. Component numbers start at 1 (for compatibility with fsl_regfilt).
In groupmelodic mode, the default output file is named “goodcomponents.txt”; components which are worth keeping will be written to MELODICDIR as integers, one per line. Component numbers start at 0 (for compatibility with standard NIFTI array indexing).
In aroma mode, the file “classified_motion_ICs.txt” must exist in the parent of MELODICDIR; by default the output will be written to “classified_motion_ICs_revised.txt” in the same directory. Component numbers start at 1 (for compatibility with AROMA numbering convention).
In fix mode, the default output file is named “hand_labels_noise.txt” and will be written to MELODICDIR as comma separated integers with square brackets surrounding the line. Component numbers start at 1 (for compatibility with FIX numbering convention).
Reprocessing fmriprep AROMA analyses
fmriprep reformats things to conform to BIDS standard naming conventions and formatting, so file locations, names, and formats are a little weird. However, you can check components as long as you used an external work directory (you set the “-w” flag during analysis).
A concrete example: I have an analysis in BIDSDIR, and used the option “-w WORKDIR” when I ran fmriprep (with AROMA processing enabled). Say I have a functional run, sub-015_ses-001_task-rest_run-1_bold.nii.gz that I want to redo the AROMA processing on. First off, I need to find my ICfile and IC mask file. They don’t get copied into the derivatives directory, as they are intermediate files, not analysis products. It turns out the entire melodic directory does exist in the work directory. In this particular case, if I set:
--melodicdir ${WORKDIR}/fmriprep_wf/single_subject_015_wf/func_preproc_ses_001_task_rest_run_1_wf/ica_aroma_wf/melodic
then PICAchooser can find the ICfile and ICmask.
The background file is also in this directory:
--backgroundfile ${WORKDIR}/fmriprep_wf/single_subject_015_wf/func_preproc_ses_001_task_rest_run_1_wf/ica_aroma_wf/melodic/mean.nii.gz
Everything else can be found in the functional output directory for this session:
FUNCDIR=${BIDSDIR}/derivatives/fmriprep/sub-015/ses-001/func
By setting the following options:
--initfile ${FUNCDIR}/sub-015_ses-001_task-rest_run-1_AROMAnoiseICs.csv --funcfile ${FUNCDIR}/sub-015_ses-001_task-rest_run-1_space-MNI152NLin6Asym_desc-preproc_bold.nii.gz --motionfile ${FUNCDIR}/sub-015_ses-001_task-rest_run-1_desc-confounds_regressors.tsv
As a bonus, if you also set:
--filteredfile ${FUNCDIR}/sub-015_ses-001_task-rest_run-1_space-MNI152NLin6Asym_desc-AROMAnonaggr_bold.nii.gz
Then when you save your bad component file, you’ll see the command necessary to refilter your data printed to the terminal window. I haven’t investigated far enough to know when the smoothing implied in the name of the exisiting filtered file comes from, so there may be some other steps to get to exactly the output you’d get from fmriprep…
melodicomp
melodicomp handles a slightly different task - its job is to allow for rapid comparison of two melodic analyses. In this case, you want to match components between the two analyses (the chances that you’ll get the same components in the same order between two analyses is basicaly zero, so you need to match them up). We do this with a spatial cross-correlation - higher cross-correlation means the the components look more like each other. This works surprisingly well. We then display the components side by side.
Usage
usage: melodicomp ICfile1 ICfile2 [options] melodicomp: error: the following arguments are required: ICfile1, ICfile2 usage: melodicomp ICfile1 ICfile2 [options]
A program to compare two sets of melodic components.
- positional arguments:
- ICfile1 The first IC component file. This will be
the exemplar, and for each component, the closest component in ICfile2 will be selected for comparison.
- ICfile2 The second IC component file. Components in
this file will be selected to match components in ICfile1.
- optional arguments:
- -h, --help
show this help message and exit
- Nonstandard input file location specification. Setting these overrides the locations assumed from ICfile1.:
- --backgroundfile BGFILE
The anatomic file on which to display the ICs (by default assumes a file called ‘mean.nii.gz’ in the same directory as ICfile1.))
- --maskfile ICMASK
The independent component mask file produced by MELODIC (by default assumes a file called ‘mask.nii.gz’ in the same directory as ICfile1.)
- --ICstatsfile1 STATSFILE
The melodic stats file (by default called ‘melodic_ICstats’ in the same directory as ICfile1),
- --ICstatsfile2 STATSFILE
The melodic stats file (by default called ‘melodic_ICstats’ in the same directory as ICfile2),
- Other arguments:
- --corrthresh CORRTHRESH
z threshold for the displayed ICA components. Default is 2.3.
- --outputfile OUTPUTFILE
Where to write the list of corresponding components (default is ‘correspondingcomponents.txt’ in the same directory as ICfile1
- --sortedfile SORTEDFILE
Save the components in ICfile2, sorted to match the components of ICfile1, in the file SORTEDFILE.
- –spatialroi XMIN XMAX YMIN YMAX ZMIN ZMAX
Only read in image data within the specified ROI. Set MAX to -1 to go to the end of that dimension.
- --displaythresh DISPLAYTHRESH
z threshold for the displayed ICA components. Default is 2.3.
- --label1 LABEL1
Label to give to file 1 components in display. Default is ‘File 1’.
- --label2 LABEL2
Label to give to file 2 components in display. Default is ‘File 2’.
- Debugging arguments:
- --verbose
Output exhaustive amounts of information about the internal workings of melodicomp. You almost certainly don’t want this.
Controls
As with PICAchooser, this is all keyboard driven. Use the right and left arrows to step through components. In melodicomp, the first file specified on the command line is considered the reference file - we go through all the components of that file and display them on the left, and show the component from the second file that matches best on the right. The number of components in the files do NOT have to match (but their spatial dimensions, voxel sizes, and background images do). Using the up and down arrows toggles between sorting based on the native order of components in file 1, and sorting in descending order of cross-correlation coefficient. Use the “a”, “c”, and “s” keys to switch between axial, coronal, and sagittal views. “b” is for blink - this swaps the right and left images. It takes essentially no time, so it makes it very clear how and where the components are changing. Try it!
By default, pairs of components with correlation coefficients lower than 0.5 are considered poor matches, and are indicated with red text in the annotations. The correlation threshold can be set on the command line.
Output
On exit (or when you hit escape), melodicomp will output a text file with the component of the first melodic file , the matching component from the second file, and the correlation coefficient between them on each line. Component numbers start from 0. The order of lines in the file is the same as the current sort order in the GUI.
rtgrader
rtgrader is my attempt at letting you quickly run through a bunch of rapidtide analyses and see which ones worked and which did not. This requires you to have a pretty good sense of what you’re looking for (which, as far as I can tell, means you have to be me, but whatever. I wrote it for me).
Usage
usage: rtgrader lagtimes strengths labels [options]
A program to review (and rate) rapidtide analyses.
- positional arguments:
- lagtimes A 4D NIFTI file of concatenated maxtime_map.nii.gz
files from a group of rapidtide analyses. These obviously all have to be at the same resolution, orentation, and coordinate system (e.g. MNI152NLin6Asm).
- strengths A 4D NIFTI file of concatenated maxcorr_map.nii.gz
files from a group of rapidtide analyses. These have to match the lagtimes file in resolution, orientation, coordinate system, and order.
- labels A text file, one entry per line, of unique descriptions
of the datasets in the lagtimes and strengths files so you know which dataset is which.
- optional arguments:
- -h, --help
show this help message and exit
- Other arguments:
- --startindex INDEX
The index to start at (0-based).
- --mapmask MASKFILE
The 3D or 4D NIFTI to mask all maps.
- –spatialroi XMIN XMAX YMIN YMAX ZMIN ZMAX
Only read in image data within the specified ROI. Set MAX to -1 to go to the end of that dimension.
- --outputroot OUTPUTROOT
Root name for reading and writing the lists of good and bad component indices (default is ‘rtgrader’, resulting in ‘rtgrader_goodindices.txt’ and ‘rtgrader_badindices.txt’)in the current working directory.
- –lagrange MINLAG MAXLAG
Lag time range to display. Default is -5.0 to 10.0.
- –strengthrange MINSTRENGTH MAXSTRENGTH
Strength range to display. Default is 0.0 to 0.75.
- Miscellaneous arguments:
- --version
show program’s version number and exit
- --detailedversion
show program’s version number and exit
- Debugging arguments:
- --verbose
Output exhaustive amounts of information about the internal workings of rtgrader. You almost certainly don’t want this.